How does DBSCAN clustering algorithm work?
the biggest secrets behind a clustering algorithm

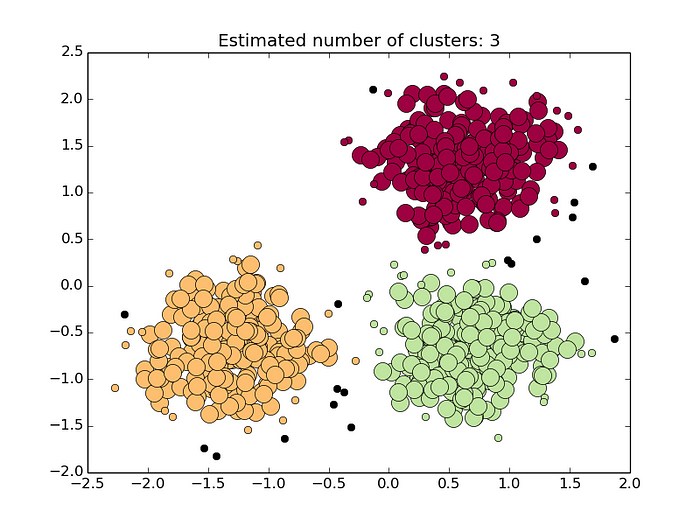
Cluster Analysis is an important problem in data analysis. And nowadays DBSCAN is one of the most popular Cluster Analysis techniques. Density-based spatial clustering of applications with noise (DBSCAN) is the data clustering algorithm proposed in the early 90s by a group of database and data mining community.
Now before we begin, let’s understand the core philosophy of DBSCAN.
Let a given dataset of points (dataset D = {xi}), we have to partition the point into the dense region which we will call them as Clusters and sparse region which may contain noise.
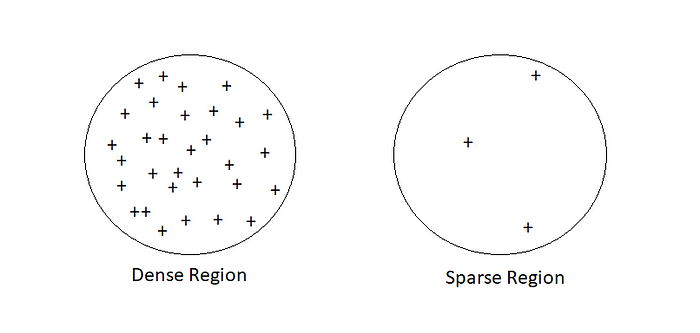
Before we can discuss density-based clustering, we first need to cover a few topics.
Parameters:
DBSCAN algorithm basically requires 2 parameters:
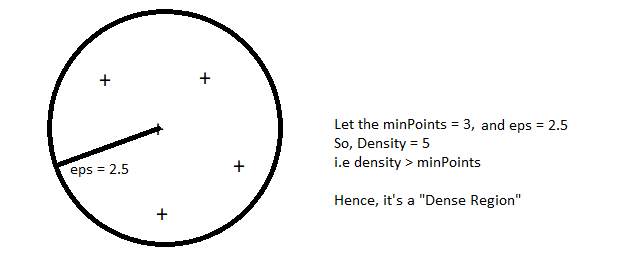
eps: specifies how close points should be to each other to be considered a part of a cluster. It means that if the distance between two points is lower or equal to this value (eps), these points are considered to be neighbors.
minPoints: the minimum number of points to form a dense region. For example, if we set the minPoints parameter as 5, then we need at least 5 points to form a dense region. It’s basically known as min_samples.
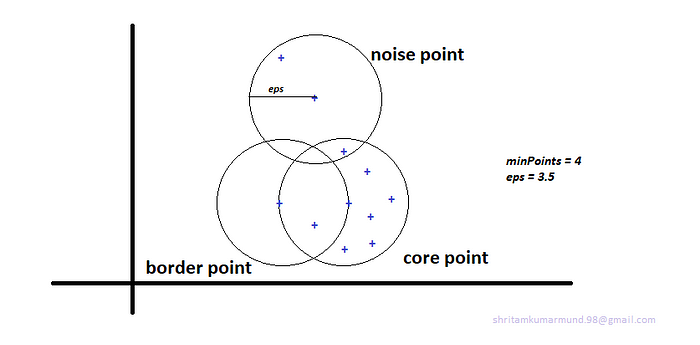
In this algorithm, we have 3 types of data points. Every point in our dataset D = {xi}, on given minPoints and eps we can categorize every data point into Core point, Border point and Noise point.
Let me define each of them.
Core point:
- A point is a core point if it has more than a specified number of minPoints within eps radius around it.
- Core Point always belongs in a dense region.
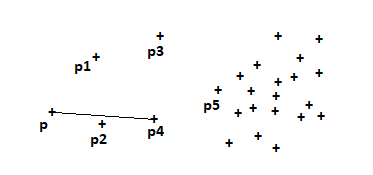
- For example, let’s consider ‘p’ is set to be a core point if ‘p’ has ≥ minPoints in an eps radius around it.
Border point:
- A point is a border point if it has fewer than MinPts within Eps, but is in the neighborhood of a core point.
- For example, p is set to be a border point if ‘p’ is not a core point. i.e ‘p’ has < minPoints in eps radius. But ‘p’ should belong to the neighborhood ‘q’. Where ‘q’ is a core point.
- p ∈ neighborhood of q and distance(p,q) ≤ eps .
Noise point:
- A noise point is any point that is not a core point or a border point.
There are two more important ideas that we should be aware of before we go and learn the DBSCAN algorithm.
Density Edge:
If p and q both are core points and distance between (p,q) ≤ eps then we can connect p, q vertex in a graph and call it “Density Edge”.
Density Connected Points:
Two points p and q are said to be density connected points if both p and q are core points and they exist a path formed by density edges connecting point (p) to point(q).

DBSCAN algorithm:
So, given all these terms and ideas, let's go into the core of the DBSCAN algorithm to see how it works.
Algorithm:
step 1: ∀ xi ∈ D, Label points as the core, border, and noise points.
step 2: Remove or Eliminate all noise points. (because they belong to the sparse region. i.e they are not belonging to any clusters.)
step 3: For every core point p that has not yet been assigned to a cluster
a) Create a new cluster with the point p and
b) add all the points that are density-connected to p.
step 4: Assign each border points to the cluster of the closest core point.
After knowing each DBSCAN concept, The question now arises:
How to choose Min Points?
- Well, one of the most important things that min Point does is to remove the outlier right!
- Typically, the rule of thumb for taking min Points is, you should always take your min points to be greater or equal to the dimensionality(d) of your dataset.
- Typically, people who work most with DBSCAN take min point twice of the dimensionality of data i.e min Point≈2*d.
- If the dataset is noisier, we should choose a larger value of min Points
- While choosing the min points, it really depends a lot on domain knowledge.
How to determine eps?
- Once you choose your min Point, you can proceed with the eps.
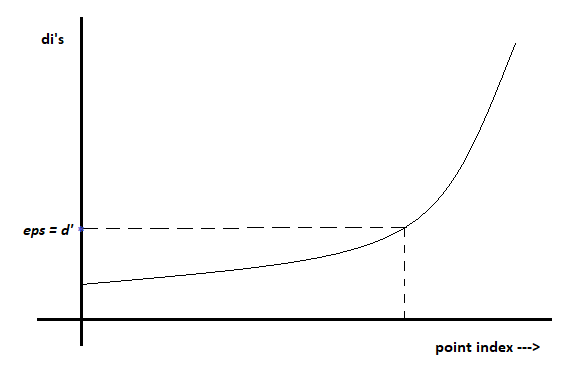
- Let us choose a min point = 4, for each point p I’ll compute “di”. where di= distance from p to the 4th nearest neighbor of p. If “di” is high, then the chance of p is noisy is also high.
- For each point in the data set, I’ll have my di’s. Now sort all di’s in increasing order.
- Since we have sorted our point in the increasing order of di’s, now we’ll plot sorted point index and di’s with elbow or Knee method.
When DBSCAN work well?
- It can handle Noise very well.
- DBSCAN can handle clusters of different shapes and sizes.
When not!
- DBSCAN, while it’s so powerful and good for some applications. There is no single algorithm that always works. Every pro has some con side.
- If your dataset has multiple densities or varying densities, DBSCAN tends to fail. It does not work very well in such cases.
- It’s extremely sensitive to the hyperparameters. A slight change in hyperparameters can lead to a drastic change in the outcome.
- As we know, a concept like Density, may not work well in high dimensionality of data. We should avoid using it for text data.
Time and Space complexity:
Time Complexity: O(n log n)
Space Complexity: O(n)
Implementation:
We don’t need to worry about implementing it ourselves. We can use one of the libraries/packages that can be found on the internet. Also, Sklearn has a DBSCAN implemented package. Let’s see how to code.
Simple Overview:
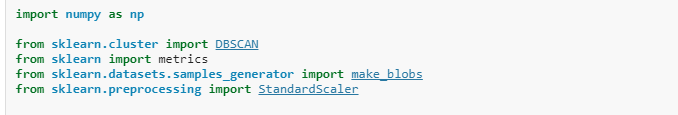
from sklearn.cluster import DBSCAN
from sklearn import metrics
import numpy as npX = #load the data
clustering = DBSCAN(eps=3, min_samples=2).fit(X)#Storing the labels formed by the DBSCAN
labels = clustering.labels_# measure the performance of dbscan algo
#Identifying which points make up our “core points”
core_samples = np.zeros_like(labels, dtype=bool)
core_samples[clustering.core_sample_indices_] = True
print(core_samples)#Calculating "the number of clusters"
n_clusters_ = len(set(labels)) - (1 if -1 in labels else 0)
print(n_clusters_)#Computing "the Silhouette Score"
print("Silhouette Coefficient: %0.3f"
% metrics.silhouette_score(X, labels))
Silhouette Coefficient Score:
The Silhouette Coefficient is calculated using the mean intra-cluster distance (
a) and the mean nearest-cluster distance (b) for each sample. The Silhouette Coefficient for a sample is(b - a) / max(a, b). To clarify,bis the distance between a sample and the nearest cluster that the sample is not a part of. Note that Silhouette Coefficient is only defined if number of labels is 2 <= n_labels <= n_samples - 1.This function returns the mean Silhouette Coefficient over all samples.
A silhouette score ranges from -1 to 1, with -1 being the worst score possible and 1 being the best score. Silhouette scores of 0 suggest overlapping clusters.
Implementation of real-world data:
So, now we are going to apply DBSCAN on the most popular real-word dataset, i.e amazon fine food review.
importing required packages
And now, your journey begins!
I hope you are now convinced to apply the DBSCAN on some datasets. It’s time to get your hands on it!
References :
Wikipedia: https://en.wikipedia.org/wiki/DBSCAN
Sklearn: https://scikit-learn.org/stable/modules/generated/sklearn.cluster.DBSCAN.html
Applied AI Course: https://www.appliedaicourse.com/
